OPTIMUS™
How does OPTIMUS™ work?
1
OPTIMUS™ is a physics-based Molecular Complexity Analysis system by BioDynLab. It leverages the Quantitative Complexity Theory (QCT) . OPTIMUS™ runs in the Scilab environment. Scilab is open-source software for numerical computation providing a powerful computing environment for engineering and scientific applications.
During execution OPTIMUS™ calls OntoNet™, a QCT computational engine, in order to process the trajectory time-histories of all atoms in a given molecule. The said time histories are processed in discrete steps via a moving-window technique. The key output of OPTIMUS™ are the Atomic Participation Factors (APF).
The APFs may be computed for single atoms in small molecules and the concept can be extended to amino acids in proteins or larger biomolecules. The APFs reflect not only the structure of a molecule but also its modes of vibration. The provide a quantitative link between structure, dynamics and biological function.
The APFs link directly to biological properties because they identify atoms whose dynamic contributions underpin functionality, stability, and interactions with biological targets (e.g., enzymes, receptors). Since biological activity (e.g., binding affinity, efficacy, toxicity) arises from dynamic processes like conformational selection or vibrational matching, atoms with high APFs are critical fragility points—modifying them amplifies or disrupts these processes, while low-APF atoms allow "safer" tweaks.
What is the input to OPTIMUS™?
2
OPTIMUS™ reads and processes PDB (Protein Data Bank) files which are produced by Molecular Dynamics Simulation codes, both commercial and free tools.
What results does OPTIMUS™ produce?
3
OPTIMUS™ produces time histories of molecular complexity, critical complexity, minimum complexity, entropy, and robustness. In addition it outputs the Atomic Participation Factors, which rank all atoms in terms of impact on a molecule’s dynamics as well as on the total encoded information. These outputs are produced in a format which makes it easy to integrate OPTIMUS™ into any SW system.
Structural robustness is particularly interesting in the case of proteins. OPTIMUS™ provides a measure of structural robustness which reflects the ability of a protein to withstand perturbations and can affect its half-life.
All of the above measures of complexity and robustness are derived deterministically.
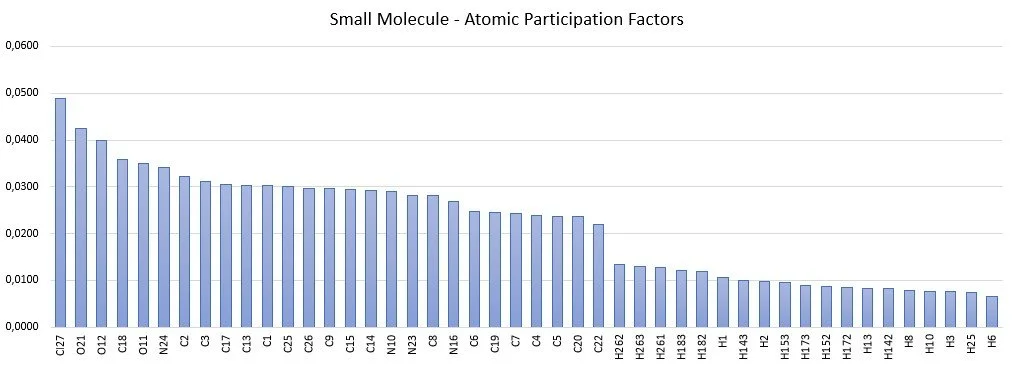
Atomic Participation Factors are intrinsic properties of a molecule. An example of Atomic Participation Factors for a small molecule is illustrated below.
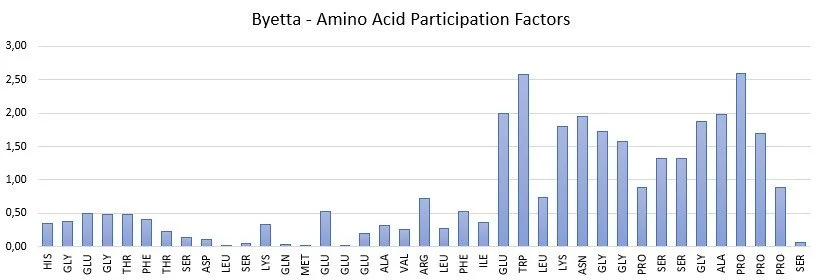
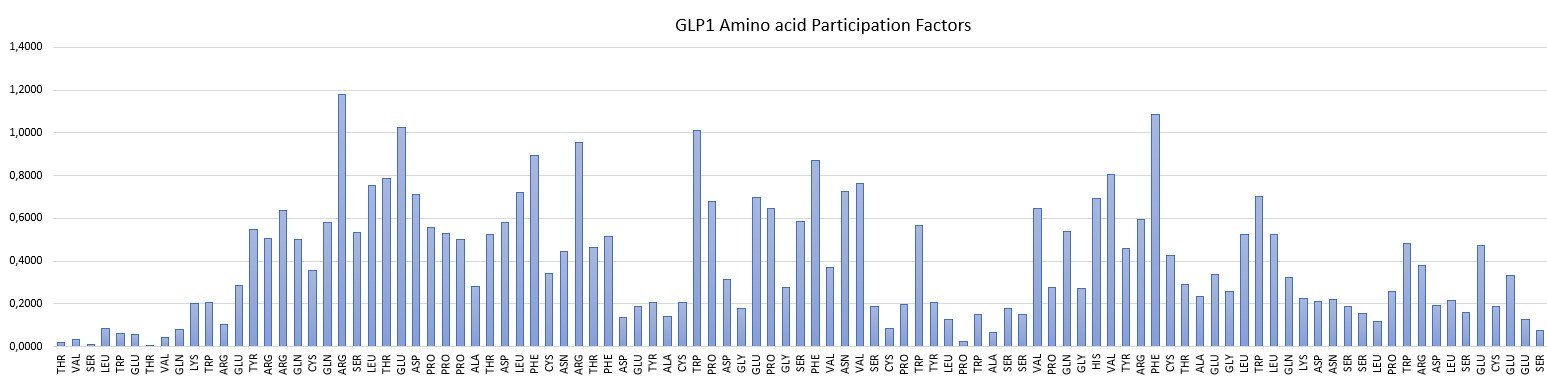
Examples of Amino acid Participation Factors for a peptide and a protein are shown below.
What is the best computational environment to run OPTIMUS™?
4
OPTIMUS™ runs best on multi-processor Linux systems. In the case of small molecules, runtime is in the order of seconds.